Data: Reticulate Evolution in Baobabs
We will be using the public data of Karimi et al, 2020 that consists of 371 aligned loci and 8 species of Adansonia plus an outgroup:
- A. digitata, continental Africa,
- A. gregorii, Australia,
- A. grandidieri, Madagascar,
- A. suarezensis, Madagascar,
- A. madagascariensis, Madagascar,
- A. perrieri, Madagascar,
- A. za, Madagascar, and
- A. rubrostipa, Madagascar.
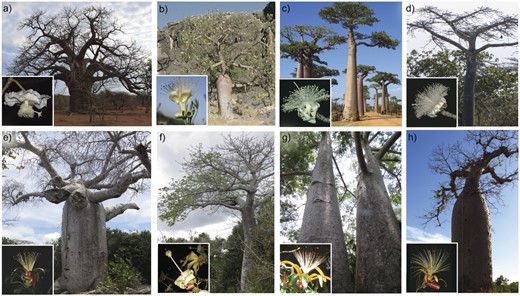
Image from Karimi et al (2020)
The original data is in FASTA format (see data/fasta-alignments), but we need NEXUS format to run MrBayes.
We have a julia script to convert FASTA to NEXUS in the repository (convertFasta2Nexus.jl), but for the sake of time, the NEXUS files are already converted in the repository (see data/nexus-alignments).
Input data: Multi-locus sequences in nexus files
We need to compress the nexus files into a tar file:
pwd ## project directory or scratch if using docker
cd snaq-tutorial/data/nexus-alignments
tar czf nexus.tar.gz *.nex
This will create a tar file (nexus.tar.gz) with the nexus files.
It is very important that you do not compress the whole folder. You should compress the individual files *.nex, not nexus-alignments or the TICR perl scripts will throw an error.
We will create a folder called analysis and move this input file there:
pwd ## inside data/nexus-alignments
cd ../..
mkdir analysis
mv data/nexus-alignments/nexus.tar.gz analysis