research
Overview

Interested in joining the lab?
Information in Opportunities.
Funding
The lab is currently funded by the following awards.
Statistical phylogenomics
In this lab, we work to produce novel statistical models and methods to reconstruct the Tree of Life that are theoretically sound yet computationally efficient and scalable to meet the ever-growing needs of biological bit data. We strive to accompany our theoretical work with open-source publicly available software.
Examples of our current research involve:
- extension of phylogenetic network inference methods to broader classes of networks
- robustness of phylogenetic inference to microbial datasets like bacteria and viruses
- alternative sampling scheme to MCMC that is expected to produce more efficient bayesian phylogenetic estimation
- statistical properties of BHV space and possible extension to networks
Our work is not purely methodological. Among our current collaborations, we can highlight:
- studying the ancestral protein sequences of Potyvirus and Picornavirus
- reconstructing phylogenetic trees and networks of grapes and carrots and studying the evolution of traits related to climate resistance via comparative methods
- reconstructing phylogenetic trees and networks of Escovopsis and studying the evolution of vesicular shapes via comparative methods
Want to learn more about phylogenetics (especially networks)? See this list of resources that starts with introductory videos and then a small subset of relevant papers in the field.
Statistics in genomics and microbiome
In this lab, we work to produce tools to better represent microbial communities (via networks) and use the microbial communities as potential predictors of plant, soil or human health phenotypes. New models are centered on high-dimensional statistical models like penalized regression and post selection inference that can simultaneously model all microbes across the microbiome.
Examples of our current research involve:
- estimation of microbial networks via Gaussian chain graph models
- network regression framework to understand the effects of microbial communities on a response
- post selection inference and penalized regression models applied to human or plant disease
- high dimensional models for the integration of different omics data types applied to human microbiome research and plant microbiome reseach
Our work is not purely methodological. Among our current collaborations, we can highlight:
- how root microbial communities affect potato health and response to environmental changes
- the effect of lung microbiome on health outcomes in cystic fibrosis patients
- the effect of gut microbiome on brain health outcomes
Statistical view in deep learning
For the last decades, deep learning has enabled unprecedented prediction potential in a plethora of applications. In particular, neural networks (NNs) are already successfully used in computer vision, astrophysics, and even in cancer histology. The main reason for their state-of-the-art accurate prediction is their flexibility, mutating its architecture to fit almost any type of data and any type of model. Yet, the poor generalization outside the training data, the lack of statistical guarantees of confidence, and the notion that they are a “black box” model have hampered their development in translational fields like personalized medicine where inaccurate predictions might result in grave consequences. Furthermore, NN methods are known for being “data-hungry”, meaning huge amounts of data are required for training and validating. This requirement prohibits its use in fields with comparatively smaller sample sizes such as human health where restrictions on data sharing and privacy limit the researchers’ ability to acquire large enough datasets for NN.
In this lab, we work to explore the potential of NN in biomedical areas. On one side, we work on data related to human health like precision medicine, or the emergence of antibiotic-resistance in infectious diseases. On the other side, we work on data related to soil and plant health like the use of biocontrol mycoviruses to fight against the emergence of fungicide-resistance crop pathogens.
Examples of our current research involve:
- robustness of NN models to predict microbial phenotypes from genomic sequences: antibiotic-resistance on Staphylococcus aureus and Pseudomonas aeruginosa and hypovirulence potential of mycoviruses on Sclerotinia
- connections of statistical concepts of uncertainty (confidence intervals or hypothesis testing) on NN models
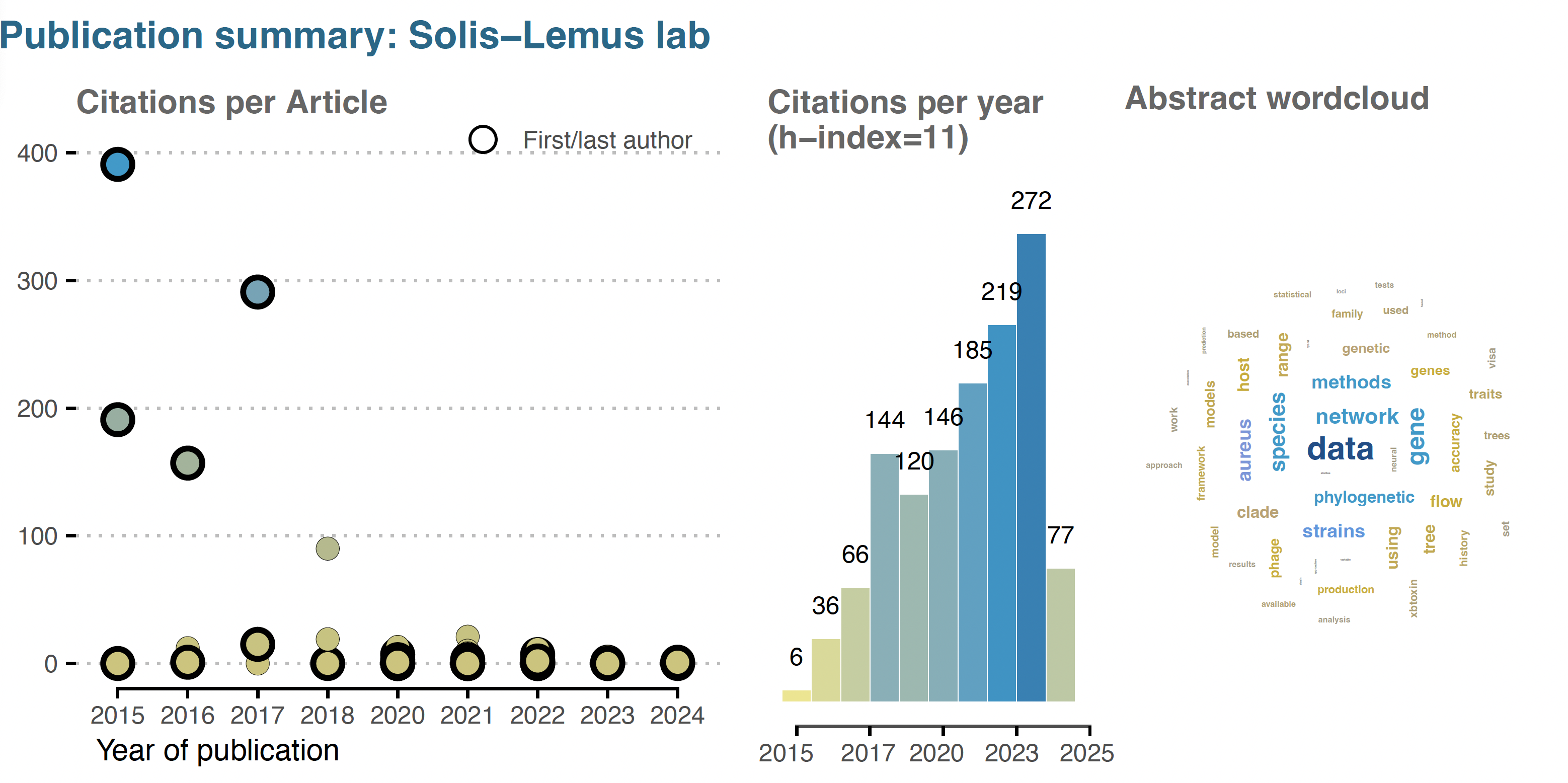
Publications
Preprints undergoing peer-review
Gorstein, E., Tang, M., Bruzzone, H.,, Solís-Lemus, C. (2026) Ancestral Sequences Cannot be Accurately Reconstructed via Interpolation in a Variational Autoencoder’s Latent Space bioarxiv:10.1101/2025.11.19.689264.
Kolbow, N., Kong, S., Chafin, T., Justison, J., Ane, C., Solís-Lemus, C. (2025) SNaQ.jl: Improved Scalability for Phylogenetic Network Inference bioarxiv:10.1101/2025.11.17.688917.
Rosas-Puchuri, U., Kolbow, N., Solís-Lemus, C., S. Khanmohammadi, R. Betancur. (2025) Sparse learning for scalable phylogenetic network inference, bioarxiv:10.1101/2025.11.16.688704.
Yang, Q., Aghdam, R., Nelson, R., Solís-Lemus, C.. (2025) MiNAA-WebApp: A Web-Based Tool for the Visualization and Analysis of Microbiome Networks, arxiv: 10.48550/arXiv:2507.08822, github, webapp
Kolbow, N., Kong, S., Solís-Lemus, C.. (2025) Massively scalable inference of level-1 phylogenetic networks, biorxiv: 10.1101/2025.05.05.652278, github
Aghdam, R., Shan, S., Lankau, R., Solís-Lemus, C.. (2025) Leveraging Bayesian Networks for Consensus Network Construction and Multi-Method Feature Selection to Decode Disease Prediction, biorxiv: 10.1101/2025.04.07.647660, github
Yang, Q., Aghdam, R., Tran, P., Anantharaman, K., Solís-Lemus, C.. (2024) Unraveling Keystone Taxa: Interactions Within Microbial Networks and Environmental Dynamics in Lake Mendota, biorxiv: 10.1101/2024.11.11.623027, github
Kong, Y., Tiley, G., Solís-Lemus, C.. (2023) Unsupervised Learning of Phylogenetic Trees via Split-Weight Embedding, arXiv:2213.16074, PhyloClustering.jl, github
Ozminkowski, S., Wu, Y., Yang, L., Xu, Z., Selberg, L., Huang, C. , Solís-Lemus, C. (2022). BioKlustering: a web app for semi-supervised learning of maximally imbalanced genomic data arxiv (2022): 2209.11730, github, bioklustering.wid.wisc.edu
Solís-Lemus, C., A. M. Holleman, A. Todor, B. Bradley, K. J. Ressler, D. Ghosh, M. P. Epstein. (2021). A Kernel Method for Dissecting Genetic Signals in Tests of High-Dimensional Phenotypes, bioRxiv 2021.07.29.454336
Shen, Y., Solís-Lemus, C. (2021). CARlasso: An R package for the estimation of sparse microbial networks with predictors, arXiv (2021): 2107.13763, github
Solís-Lemus, C., A. Coen and Cecile Ané. 2020. On the identifiability of phylogenetic networks under a pseudolikelihood model, arxiv (2020): 2010.01758, github
| 2026 | Shen, Y., Solís-Lemus, C. | |
| Bayesian chain graph models to characterize microbe-environment dynamics | ||
| DOI: 10.3934/mbe.2026020 | ||
| 2025 | Aghdam, R., Solís-Lemus, C. | |
| CMiNet: R package for learning the Consensus Microbiome Network | ||
| DOI: 10.1111/2041-210x.70198 | ||
| 2025 | Tiley, G., Liu, N., Solís-Lemus, C. | |
| Extracting diamonds: Identifiability of 4-node cycles in level-1 phylogenetic networks | ||
| DOI: 10.1093/evolinnean/kzaf019 | ||
| 2025 | Sedlacek, Q. C., Friend, M., Villa, A. M. V., Dozier, S., León, M., Haeger, H., Lomelí, K., Pérez, G., Solís-Lemus, C., & Mejia, J. A. | |
| STEM disciplines are more diverse than undergraduate courses depict | ||
| DOI: 10.1007/s11191-025-00701-9 | ||
| 2025 | Q. Vidaurre Montoya, N. M. Gerardo, M. J. S. Martiarena, Solís-Lemus, C., R. Kriebel, T. R. Schultz, J. Sosa-Calvo & A. Rodrigues | |
| Digging into the evolutionary history of the fungus-growing-ant symbiont, Escovopsis (Hypocreaceae) | ||
| DOI: 10.1038/s42003-025-08654-z | ||
| 2025 | Kong, S., Solís-Lemus, C. and Tiley, G. | |
| Phylogenetic networks empower biodiversity research | ||
| DOI: 10.1073/pnas.2410934122 | ||
| 2025 | Peñaloza-Bojacá, Burleigh, Maciel-Silva, Cargill, Bell, Sessa, McDaniel, Davis, Endara, Salazar Allen, Li, Schafran, Chantanaorrapint, Duckett, Pressel, Solís-Lemus, C., Renzaglia, Villarreal | |
| Ancient reticulation, incomplete lineage sorting and the evolution of the pyrenoid at the dawn of hornwort diversification | ||
| DOI: 10.1093/aob/mcaf002 | ||
| 2025 | Solís-Lemus, C. and Tiley, G. | |
| Seeing the Network for the Trees: Methodological and Empirical Advances in Reticulate Evolution | ||
| DOI: 10.18061/bssb.v4i1.10445 | ||
| 2025 | Gorstein, E., Aghdam, R., Solís-Lemus, C. | |
| HighDimMixedModels.jl: Robust High Dimensional Mixed Models across Omics Data | ||
| DOI: 10.1371/journal.pcbi.1012143 | ||
| 2024 | Aghdam, R., Tang., X., Shan, S., Lankau, R., Solís-Lemus, C. | |
| Human Limits in Machine Learning: Prediction of Plant Phenotypes Using Soil Microbiome Data. | ||
| DOI: 10.1186/s12859-024-05977-2 | ||
| 2024 | Bjorner, M., Molloy, E., Dewey, C., Solís-Lemus, C. | |
| Detectability of Varied Hybridization Scenarios Using Genome-Scale Hybrid Detection Methods | ||
| DOI: 10.18061/bssb.v3i1.9284 | ||
| 2024 | Rosas-Puchuri, U., Santaquiteria, A., Khanmohammadi, S., Solís-Lemus, C., Betancur, R. | |
| Non-linear phylogenetic regression using regularised kernels | ||
| DOI: 10.1111/2041-210X.14385 | ||
| 2024 | Tiley, George P., Andrew A. Crowl, Paul S. Manos, Emily B. Sessa, Solís-Lemus, C., Anne D. Yoder, and J. Gordon Burleigh | |
| Benefits and Limits of Phasing Alleles for Network Inference of Allopolyploid Complexes | ||
| DOI: 10.1093/sysbio/syae024 | ||
| 2024 | Ozminkowski, S., Solís-Lemus, C. | ||
| Identifying microbial drivers in biological phenotypes with a Bayesian Network Regression model | |||
| DOI: 10.1002/ece3.11039 | |||
| 2024 | Shen, Y., Solís-Lemus, C. | ||
| The Effect of the Prior and the Experimental Design on the Inference of the Precision Matrix in Gaussian Chain Graph Models | |||
| DOI: 10.1007/s13253-024-00621-1 | |||
| 2024 | Nelson, R., Aghdam, R., Solís-Lemus, C. | ||
| MiNAA: Microbiome Network Alignment Algorithm | |||
| DOI: 10.2115/joss.05448 | |||
| 2024 | Shen, Y., Solís-Lemus, C., Deshpande, S.K. | |
| Estimating sparse direct effects in multivariate regression with the spike-and-slab LASSO | ||
| DOI: 10.1214/24-BA1430 | ||
| 2024 | Wu, Z., Solís-Lemus, C. | ||
| Ultrafast learning of 4-node hybridization cycles in phylogenetic networks using algebraic invariants | |||
| DOI: 10.1093/bioadv/vbae014 | |||
| 2024 | Tang, X., L. Zepeda-Nunez, S. Yang, Z. Zhao, Solís-Lemus, C. | ||
| Novel Symmetry-preserving Neural Network Model for Phylogenetic Inference | |||
| DOI: 10.1093/bioadv/vbae022 | |||
| 2023 | Rattray, JB, Walden, R., Marquez-Zacarias, P., Molotkova, E., Perron, G., Solís-Lemus, C., Pimentel-Alarcon, D., Brown, S. | ||
| Machine learning identification of Pseudomonas aeruginosa strains from colony image data. | |||
| DOI: 10.1371/journal.pcbi.1011699 | |||
| 2023 | Justison, J., Solís-Lemus, C., Heath, T. | ||
| SiPhyNetwork: An R package for simulating phylogenetic networks | |||
| DOI: 10.1111/2041-210X.14116 | |||
| 2022 | Sun, Y., T.M. Maeda, Solís-Lemus, C., D. Pimentel-Alarcon, Z. Burivalova | ||
| Classification of animal sounds in a hyperdiverse rainforest using Convolutional Neural Networks | |||
| DOI: 10.1016/j.ecolind.2022.109621 | |||
| 2022 | Liu, Y., Solís-Lemus, C. | ||
| WI Fast Stats: a collection of web apps for the visualization and analysis of WI Fast Plants data | |||
| DOI: 10.21105/jose.00159 | |||
| 2022 | Zhang, Z., Cheng, S., Solís-Lemus, C. | |
| Towards a robust out-of-the-box neural network model for genomic data | ||
| DOI: 10.1186/s12859-022-04660-8 | ||
| 2022 | G. A. Satten, S. W. Curtis, C. Solís-Lemus, C., E. J. Leslie, M. P. Epstein | |
| Efficient Estimation of Indirect Effects in Case-Control Studies Using a Unified Likelihood Framework | ||
| DOI: 10.1002/sim.9390 | ||
| 2021 | Su M, Davis MH, Peterson J, Solís-Lemus, C., Satola SW, Read TD. | |
| Effect of genetic background on the evolution of Vancomycin-Intermediate Staphylococcus aureus (VISA) | ||
| DOI: 10.7717/peerj.11764 | ||
| 2021 | Moller A., Winston K., Ji S., Wang J., Hargita Davis M., Solís-Lemus, C., Read T. | |
| Genes Influencing Phage Host Range in Staphylococcus aureus on a Species-Wide Scale | ||
| DOI: 10.1128/mSphere.01263-20 | ||
| 2020 | Guerrero, V. and Solís-Lemus, C. | |
| A generalized measure of relative dispersion | ||
| DOI: 10.1016/j.spl.2020.108806 | ||
| 2020 | Solís-Lemus, C., S. T. Fischer, A. Todor, C. Liu, E. J. Leslie, D. J. Cutler, D. Ghosh and M. P. Epstein | |
| Leveraging Family History in Case-Control Analyses of Rare Variation | ||
| DOI: 10.1534/genetics.119.302846 | ||
| 2020 | M. Su, J. Lyles, R. A. Petit III, J. M. Peterson, M. Hargita, H .Tang, Solís-Lemus, C., C. Quave, T. D. Read | |
| Genomic analysis of variability in delta-toxin levels between Staphylococcus aureus strains | ||
| DOI: 10.7717/peerj.8717 | ||
| 2020 | Solís-Lemus, C., Ma, X., Hostetter II, M., Kundu, S., Qiu, P., Pimentel-Alarcón D. | |
| Prediction of functional markers of mass cytometry data via deep learning | ||
| DOI: 10.1007/978-3-030-33416-1_5 | ||
| 2018 | Solís-Lemus, C., Pimentel-Alarcón D. | |
| Breaking the Limits of Subspace Inference | ||
| 56th Annual Allerton Conference on Communication, Control, and Computing |
| 2018 | Spooner, D.M., Ruess, H., Arbizu, C.I., Rodriguez, F., Solís-Lemus, C. | |
| Greatly reduced phylogenetic structure in the cultivated potato clade (Solanum section Petota pro parte) | ||
| DOI: 10.1002/ajb2.1008 | ||
| 2018 | Bastide, P., Solís-Lemus, C., Kriebel, R., Sparks, K.W., Ané, C. | |
| Phylogenetic Comparative Methods on Phylogenetic Networks with Reticulations | ||
| DOI: 10.1093/sysbio/syy033 | ||
| 2017 | Solís-Lemus, C., Bastide, P., Ané, C. | |
| PhyloNetworks: a package for phylogenetic networks | ||
| DOI: 10.1093/molbev/msx235 | ||
| 2017 | Ané, C., Bastide, P., Mariadassou, M., Robin, S., Solís-Lemus, C. | |
| Processus d’évolution réticulée: tests de signal phylogénétique | ||
| Journées de Statistique |
| 2017 | Pimentel-Alarcón D., Biswas A., Solís-Lemus, C. | |
| Adversarial Principal Component Analysis | ||
| IEEE International Symposium on Information Theory (ISIT) |
| 2016 | Solís-Lemus, C., Ané, C. | |
| Inferring phylogenetic networks with maximum pseudolikelihood under incomplete lineage sorting | ||
| DOI: 10.1371/journal.pgen.1005896 | ||
| 2016 | Solís-Lemus, C., Yang, M., Ané, C. | |
| Inconsistency of species-tree methods under gene flow | ||
| DOI: 10.1093/sysbio/syw030 | ||
| 2016 | Baum, D., Ané, C., Larget, B., Solís-Lemus, C., Ho, L.S.T, Boone, P., Drummond, C., Bontrager, M., Hunter, S., Saucier, B. | |
| Statistical evidence for common ancestry: application to primates | ||
| DOI: 10.1111/evo.12934 | ||
| 2016 | Pimentel-Alarcón D., Solís-Lemus, C. | |
| Crime detection via crowdsourcing | ||
| 8th Mexican Conference on Pattern Recognition, Springer International |
| 2015 | Solís-Lemus, C., L.L. Knowles and C. Ané | |
| Bayesian species delimitation combining multiple genes and traits in a unified framework | ||
| DOI: 10.1111/evo.12582 | ||
| 2015 | Solís-Lemus, C. | |
| Statistical methods to infer population structure with coalescence and gene flow. | ||
| PhD dissertation, Department of Statistics, University of Wisconsin-Madison | ||
Awards
NSF CAREER
Title: CAREER: Towards Scalable and Robust Inference of Phylogenetic Networks
Dates: February 1, 2022 to January 31, 2027
Personnel:
- PI: Claudia Solis-Lemus
- Ben Rush (postdoc)
- Sungsik Kong (postdoc)
- Rosa Aghdam (postdoc)
- Marianne Bjorner (MS student)
- Xudong Tang (MS student)
- Hailey Louw (MS student)
- Nathan Kolbow (PhD student)
- Evan Gorstein (PhD student)
- Jiayang Wang (PhD student)
- Yuke Wu (undergrad)
- Bella Wu (undergrad)
- Yibo Kong (undergrad)
- Lakes Tang (undergrad)
Project summary
Scientists world-wide are engaged in efforts to understand how all planetary biodiversity evolved. This diversification process is represented through the Tree of Life. Achieving the goal of a complete estimate of the Tree of Life would allow us to fully understand the development and evolution of important biological traits in nature, for example, those related to resilience to extinction when exposed to environmental threats such as climate change. It would also provide information about the emergence and evolution of novel human pathogens that pose severe threats to human health. Thus, the development of statistical and computational tools to reconstruct the Tree of Life are paramount in evolutionary biology, systematics, conservation efforts, and human health research. Existing tree reconstruction methods, however, are limited because they do not account for important biological processes such as species hybridization, introgression or horizontal gene transfer, and thus, recent years have seen an explosion of methods to reconstruct phylogenetic networks rather than trees. Existing network reconstruction methods lack statistical guarantees ensuring the detection of reticulate signals in data, are not scalable enough for big data, and are tailored to reconstruct simple networks. Thus, they are not sufficient to tackle the complexity of reticulate evolution in fungi, prokaryotes, or viruses. This project will develop novel network inference methods with strong statistical guarantees that are robust enough to infer complex networks and scalable enough to accommodate big data. The methods will allow the integration of all organisms into the Tree of Life and thus help to complete a broader picture of evolution across all domains of life. The project will produce open source software and data science modules for K-16 outreach, and includes a strong focus on training underrepresented groups in STEM.
Publications supported by the award
- Kong et al (2023). arXiv:2213.16074
- Tiley and Solis-Lemus (2023). biorXiv:2023/564087
- Justison et al (2023). MEE DOI: 10.1111/2041-210X.14116
- Wu and Solis-Lemus (2024). BioAdv DOI: 10.1093/bioadv/vbae014
- Tang et al (2024). BioAdv DOI: 10.1093/bioadv/vbae022
- Tiley et al (2024). Syst Bio DOI: 10.1093/sysbio/syae024
- Aghdam and Solis-Lemus (2024). arXiv:2411.08309
- Yang et al (2024). biorxiv: 10.1101/2024.11.11.623027
- Bjorner et al (2024). DOI: 10.18061/bssb.v3i1.9284
- Gorstein et al (2025). DOI: 10.1371/journal.pcbi.1012143
- Aghdam et al (2025). biorxiv: 10.1101/2025.04.07.647660
- Solis-Lemus and Tiley (2025). DOI: 10.18061/bssb.v4i1.10445
- Kolbow et al (2025). biorxiv: 10.1101/2025.05.05.652278
- Yang et al (2025). arxiv: 10.48550/arXiv:2507.08822
- Kong et al (2025). DOI: 10.1073/pnas.2410934122
- Tiley et al (2025). DOI: 10.1093/evolinnean/kzaf019
- Aghdam and Solis-Lemus (2025). DOI: 10.1111/2041-210x.70198
- Rosas-Puchuri et al (2025). bioarxiv:10.1101/2025.11.16.688704
- Kolbow et al (2025). bioarxiv:10.1101/2025.11.17.688917
USDA Individual Hatch
Project summary
The growing food demand can only be sustained through rigorous and consistent sup- port of plant and soil health worldwide. Recognizing the microbial, environmental and agricultural factors that drive plant and soil phenotypes is crucial to comprehend processes connected to plant and soil health, to identify global practices of sustainable agriculture, as well as to predict plant and soil responses to environmental perturbations such as climate change. In order to identify the driving factors in plant and soil health, we need robust statistical tools that are able to connect a set of predictors with a specific phenotype. Yet, the innovation in the methodological data science tools for agricultural practices has not matched the increasing complexity of soil and plant data.
The overall objective of this project is to develop a next generation of statistical theory (accompanied by open-source publicly available software) for soil and plant high-dimensional data. Our novel statistical methods will overcome existing challenges in standard approaches in two ways: 1) they will inherently account for high-dimensional highly interconnected data through the development of novel microbiome interaction models, and 2) they will explore the inclusion of graphical predictors such as microbiome interaction networks through the theory of network regression analysis.
Publications supported by the award
- Shen and Solis-Lemus (2026) DOI: 10.3934/mbe.2026020
DOE: Computational Tool Development for Integrative Systems Biology Data Analysis
Title: Harnessing the power of big omics data: Novel statistical tools to study the role of microbial communities in fundamental biological processes
Dates: September 14, 2020 to September 14, 2022
Personnel:
- PI: Claudia Solis-Lemus
- Sam Ozminkowski (MS student)
- Marianne Bjorner (MS student)
- Rosa Aghdam (postdoc)
- Yuke Wu (undergrad)
- Reed Nelson (undergrad)
Project summary
Microbial communities are among the main driving forces of biogeochemical processes in the biosphere. In particular, many critical soil processes such as mineral weathering, and soil cycling of mineral-sorbed organic matter are governed by mineral-associated microbes. Understanding the composition of microbial communities and what environmental factors play a role in shaping this composition is crucial to comprehend soil biological processes and to predict microbial responses to environmental changes. In order to identify the driving factors in soil biological processes, we need robust statistical tools that are able to connect a set of predictors with a specific phenotype. Yet, the innovation in the statistical theory for biochemical and biophysical processes has not matched the increasing complexity of soil data. Indeed, existing statistical techniques have four main drawbacks: 1) they perform poorly on high-dimensional highly sparse data, such as soil metagenomics; 2) they ignore spatial correlation structure which can be a key component in soil-related data; 3) they do not provide valid p-values under high-dimensional settings making them unable to detect significant factors driving the phenotype of interest, and 4) they tend to focus on abundance matrices to represent microbial compositions which can be flawed due to its compositional nature (sum to 1 restriction) that affects how proportions behave in different experimental settings (e.g. changes in proportions in the microbial composition does not necessarily reflect actual biological changes in the interactions). The overall objective of this proposal is to pioneer the development of the next generation of statistical theory (accompanied by open-source publicly available software) for soil omics data. Our novel statistical methods will overcome existing challenges in standard approaches in three ways: 1) they will inherently account for high-dimensional highly interconnected data through the development of novel mixed-effects sparse learning models; 2) they will produce valid adaptive p-values through post selection inference, and 3) they will be implemented in open-source publicly available software that will serve the broader scientific community.
Publications supported by the award
- Zhang et al (2020) BMC Bioinformatics DOI:10.1186/s12859-022-04660-8
- Liu and Solis-Lemus (2020) JOSE DOI:10.21105/jose.00159
- Shen and Solis-Lemus (2021) arXiv:2107.13763
- Ozminkowski et al (2022) arxiv:2209.11730
- Tang et al (2024) BioAdv DOI: 10.1093/bioadv/vbae022
- Shen et al (2024) BA DOI: 10.1214/24-BA1430
- Nelson et al (2024) JOSS DOI: 10.2115/joss.05448
- Ozminkowski and Solis-Lemus, C. (2024) EcoEvo DOI: 10.1002/ece3.11039
- Shen and Solis-Lemus (2024) JABES DOI: 10.1007/s13253-024-00621-1
- Bjorner et al (2024). DOI: 10.18061/bssb.v3i1.9284
- Aghdam et al (2024) DOI: 10.1371/journal.pcbi.1012143
- Gorstein et al (2025). DOI: 10.1371/journal.pcbi.1012143
- Shen and Solis-Lemus (2026) DOI: 10.3934/mbe.2026020
USDA-NIFA: hatch project 1023699
Project summary
The growing food demand can only be sustained through rigorous and consistent support of plant and soil health worldwide. Recognizing the microbial, environmental and agricultural factors that drive plant and soil phenotypes is crucial to comprehend processes connected to plant and soil health, to identify global practices of sustainable agriculture, as well as to predict plant and soil responses to environmental perturbations such as climate change. In order to identify the driving factors in plant and soil health, we need robust statistical tools that are able to connect a set of predictors with a specific phenotype. Yet, the innovation in the methodological data science tools for agricultural practices has not matched the increasing complexity of soil and plant data. The overall objective of this project is to develop a next generation of statistical theory (accompanied by open-source publicly available software) for soil and plant data by exploiting the high- dimensional highly interconnected data through the development of novel microbiome interaction models. By harnessing the power of big data through new statistical theory in sparse learning, and network regression models, our work will produce tools that can better understand the drivers in soil and plant health to aid in the adoption of global practices of sustainable agriculture, which are vital to meet the ever-increasing need for food availability in the XXI century.
Publications supported by the award
- Shen and Solis-Lemus (2021) arXiv:2107.13763
- Ozminkowski et al (2022) arxiv (2022): 2209.11730
- Shen et al (2024) BA DOI: 10.1214/24-BA1430
- Ozminkowski and Solis-Lemus, C. (2024) EcoEvo DOI: 10.1002/ece3.11039
- Shen and Solis-Lemus (2024) JABES DOI: 10.1007/s13253-024-00621-1
- Shen and Solis-Lemus (2026) DOI: 10.3934/mbe.2026020
Wisconsin Potato and Vegetable Growers Association, Inc.
Project summary
The overarching objective of this proposal is to initiate development of a virtual tool for analyzing and visualizing field data collected each year by the Wisconsin Seed Potato Certification Program (WSPCP) for use on the plant health certificate. Specific objectives include: 1) Creating an enhanced cloud-based database to house seed certification program data, 2) Developing visualization tools for interacting with seed potato certification program data, and 3) Generating data analytics capability to extrapolate from trends in the available data.
Software supported by the award
- Potato Dashboard (only available for WI seed certification staff at the moment)
Collaborative Awards
NSF IntBIO Collaborative Research
Title: IntBIO Collaborative: Assessing drivers of the nitrogen-fixing symbiosis at continental scales
Dates: August 1, 2023 to July 31, 2027
Lead PI: Ryan Folk, Mississippi State U
Personnel:
- co-PI: Claudia Solis-Lemus
Project summary
Interactions between plants and bacteria are pervasive, and nitrogen-fixing symbioses are among the most important of these given the generality of nitrogen limitation and the inability of eukaryotes to access atmospheric nitrogen directly. Despite a long history of interest and the clear ecological importance and agricultural potential of nitrogen-fixing symbiosis, still missing is a cohesive framework for understanding the joint action of ecological and evolutionary processes that shape this symbiosis. We therefore lack a clear conceptual basis for how the environment, plants, and microbes interact at local scales to produce a successful symbiosis across diverse environmental and biogeographic contexts. We propose to build such a framework by gathering extensive data on plant-microbe interactions that occur from the soil to within root nodules, and assessing how the intimacy of these interactions depends on geographic, environmental, and phylogenetic scales. The proposed research will integrate with existing ecological monitoring resources at NEON (National Ecological Observatory Network) sites to capture microbes for the majority of nodulating nitrogen-fixing plant species, investigating microbial (fungal and bacterial) diversity along four sampling levels: soils, the rhizosphere, the nodule, and non-nodular roots. Our hypothesis-based work has three broad goals: (1) assess the importance of environmental drivers, extending breaking results showing that aridity and secondarily soil nutrients determine nitrogen-fixing plant diversity and asking whether nitrogen-fixing and other bacterial and fungal symbionts are constrained by similar processes as the host; (2) characterize the host-specificity of nodulators and their symbionts at broad and narrow phylogenetic scales, asking whether bacterial-host match-up determines downstream functional symbiosis efficiency or whether instead particular symbionts are dispensable; and (3) test for local-scale and deep-level co-phylogeny of nodulating plants and their bacterial partners, and whether the strength of co-phylogeny depends on environmental contingencies of the need for symbiosis.
Publications supported by the award
- [upcoming]
USDA NIFA
Project summary
The primary goals of this proposal are to assess how fungicide applications disturb soil and foliar plant microbiomes and determine the subsequent impacts of this microbial dysbiosis on plant disease development. Preliminary data collected in turfgrass by PD Koch and co-PI Chou with the fungal disease dollar spot (caused by Clarireedia jacksonii) revealed a dramatic increase in dollar spot severity in the weeks following repeated applications of the broad-spectrum fungicide chlorothalonil. This ‘disease resurgence’ occurred after the pathogen-suppressive activity of the fungicide had ended, and dollar spot severity in the chlorothalonil-treated plots was six times higher than non-treated control plots that had not received any fungicide that season. Disease resurgence was not observed after applications of the fungicide propiconazole, which has a more targeted spectrum of microbial suppression compared to chlorothalonil. Initial microbiome characterization from that preliminary study found some indications of microbial dysbiosis, but we are proposing here a more robust soil and foliar microbial community and metabolomic analysis to explore potential microbial mechanisms mediating disease resurgence in both turfgrass and corn.
Publications supported by the award
- [upcoming]
NSF ADVISE
Project summary
The Amplifying Diverse Voices in STEM Education (ADVISE) project will use a cluster randomized experiment across four HSIs and five PWIs to test the causal effects of bringing racially marginalized guest lecturers into college STEM courses. The overarching goal of this collaborative project is to better understand and compare individual- and group-level psychological and sociological processes such as student belonging, stereotype threat, classroom climate, and instructor pedagogy, and how guest lectures may affect these processes in ways that can promote equitable and effective STEM education. Outcomes, mediators, and moderators will be measured using repeated-measures surveys and FERPA-compliant institutional data on student course grades and persistence in STEM majors. Internal and external project evaluation will use a collective impact model to ensure effective, equity-focused STEM education research. Findings will be disseminated through academic presentations and publications as well as reports and publications for policymaker and practitioner audiences in higher education and STEM fields. Beyond basic science contributions to social psychology and sociology and applied science contributions to higher education and STEM education, broader impacts are expected to include improved experiences and outcomes for thousands of college STEM students, professional opportunities for hundreds of early-career STEM scholars, and diversified professional networks for current STEM faculty.
Publications supported by the award
- [upcoming]



