teaching
Courses at UW-Madison
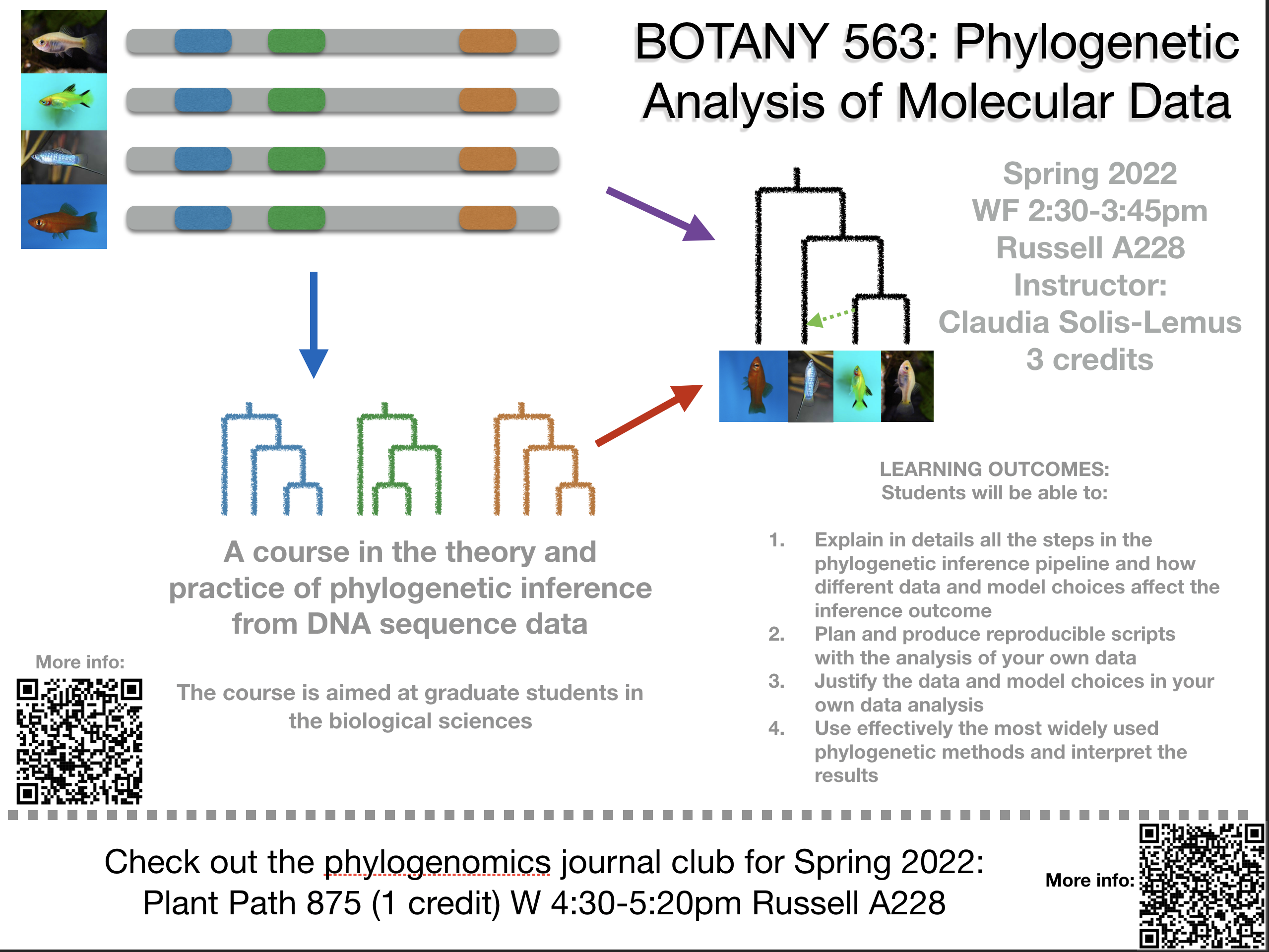
Botany/PlantPath 563 Phylogenetic Analysis of Molecular Data
- Spring 2021: 3 credits; TR 1:00-2:15pm (virtual over zoom)
- Spring 2022: 3 credits; WF 2:30-3:45pm
- Spring 2023: 3 credits; TR 1:00-2:15pm
- Spring 2024: 3 credits; TR 1:00-2:15pm
- Spring 2025: 3 credits; TR 1:00-2:15pm
- Spring 2026: 3 credits; TR 1:00-2:15pm
Plant Path 875 Readings in Phylogenomics
- Spring 2021: 1 credit; F 8:55-9:45am (virtual over zoom)
- Spring 2022: 1 credit; W 4:30-5:20pm
Plant Path 875 Phylogenetics Practicum
- Spring 2026: 1 credit; F 10:00-10:50am
Workshops
Spring 2025
- Forensic Justice Institute 2025 by the State Bar of Wisconsin
Fall 2024
Spring 2024
- Methods for Biological Data Workshop
- Forensic Justice Institute 2024 by the State Bar of Wisconsin
Fall 2023
- Methodological advances in reticulate evolution Workshop co-organized by George Tiley in Kew Botanical Garden, UK:

Summer 2023
- Taming the BEAST Faculty member of the workshop on BEAST. Lecture on phylogenetic networks (taught by postdoc Sungsik Kong). Squamish, Canada.

- Botany conference Workshop on phylogenetic networks. Boise, Idaho.
- Molecular Evolution in MBL: Lecture and tutorial on phylogenetic networks. Woods Hole, USA
Spring 2023
- DNA Bootcamp 2023 by California State University Los Angeles, Los Angeles Innocence Project, Federal Defender Central District of California, and Los Angeles County Public Defender
- Accessible reproducibility for biological researchers at the Toronto Workshop on Reproducibility
- SSB Satellite Mexico City: Introduction to PhyloNetworks.
Summer 2022
- AI+Science Summer school: Deep learning in biology. University of Chicago. August 10, 2022.
-
Woods Hole: Faculty member of the Marine Biological Laboratory workshop on Molecular Evolution. Lecture and tutorial on phylogenetic networks. Woods Hole, USA.
- Julia workshop for Data Science co-organized with Doug Bates for ISMB 2022 on July 10th, 2022. Website, GitHub
- Statistics for Forensics 101 (National Forensic College): website
Spring 2022
- Introduction to Statistics for Defense Attorneys (WI public defenders): website
Fall 2021
BME 780 Guest lecture: Statistical phylogenomics
- Optional background reading:
- Lecture notes (Oct 26): slides
- Paper discussion (Nov 3): Multispecies coalescent and its applications to infer species phylogenies and cross-species gene flow
- Learn more about phylogenomics:
- HAL open access book Chapters 1.1, 1.2, 1.4, 3.1 and 3.3
- Phylogenetic Trees and Networks Can Serve as Powerful and Complementary Approaches for Analysis of Genomic Data
- Spring 2022 Botany 563 Phylogenetic Analysis of Molecular Data
STAT 609/849 Guest lecture: Statistical methods for biological applications (Oct 19)
Summer 2021
Virtual species delimitation workshop at the National Museum of Natural History (NMNH); August 16-19, 2021. iBPP wiki, YouTube video
Spring 2021
Julia para Ciencias de Datos organized by Seminario de Investigacion de la Escuela de Estadistica de la Universidad de Los Andes, Merida, Venezuela. Github
Julia workshop for Data Science co-organized with Doug Bates for the 2021 Data Science Research Bazaar on February 10th, 2021. GitHub
Fall 2020
Mexicanas en Ciencias de Datos: Julia workshop for Data Science. github
STAT 609/849 Guest lecture: Statistical methods for biological applications (Oct 1) ![]()
2019
Nantucket developR: Tutorial on best computing practices using R: mindful programming. Notes from other instructors here.
Taming the BEAST Faculty member of the workshop on BEAST. Lecture on phylogenetic networks. Squamish, Canada

Woods Hole: Faculty member of the Marine Biological Laboratory workshop on Molecular Evolution. Lecture and tutorial on phylogenetic networks. Woods Hole, USA.
2016:
Botany 563: Guest lecture on Introduction to phylogenetic networks
Instituto de Biología - UNAM: Introduction to PhyloNetworks and SNaQ: Same tutorial as in Evolution 2016, links below.
Evolution: Phylogenomics symposium and software school: Introduction to PhyloNetworks and SNaQ
Useful links
Reprodubility, computing practices and open science
- My notes on mindful programming and accompanying YouTube video
- Karl Broman’s notes on reproducible research
- Version control: notes from Software Carpentry at UW-Madison (there might be more recent notes out there)
- R Markdown: The Definite Guide
- Rich Edwards (2015) Bioinformatics is just like bench science and should be treated as such
- Basic principles of data in spreadsheets paper
- JSM 2020 Panel on version control with git in stat cources
- Bioinformatics one-liners
Learning Julia
- Learn X in Y minutes: julia
- Julia for R programmmers: cheatsheet
- Julia for Python/Matlab programmers: cheatsheet
- Julia for Data Science workshop
- How to create a Julia package
Learning Statistics
- Tukey (1962) The future of data analysis
- StatQuest: YouTube channel with short videos to explain important concepts in Statistics and Data Science
Other teaching experience
Teaching Assistant (UW-Madison)
- Summer 2014 PhD qualifier Summer camp
- Spring 2013 Introduction to Biostatistics for Population Health II (BIOSTAT 552) (professor Ron Gangnon)
- Fall 2012 Introduction to Biostatistics for Population Health I (BIOSTAT 551) (professor Ron Gangnon)
- Spring 2012 Introduction to Biostatistics for Population Health II (BIOSTAT 552) (professor Ron Gangnon)
- Fall 2011 Introduction to Biostatistics for Population Health I (BIOSTAT 551) (professor Ron Gangnon)
- Summer 2011 Introduction to Statistical Methods (STAT 301) (professor Kam Wah Tsui)
- Spring 2011 Introduction to Statistical Methods (STAT 301) (professor Kevin Packard)
- Fall 2010 Introduction to Theory and Methods of Mathematical Statistics I (STAT 311) (professor Zhengjun Zhang)
Instructor (ITAM)
- Summer 2010 Advanced Algebra
- Spring 2010 Calculus II
- Fall 2009 Calculus III
- Fall 2009 Actuarial Mathematics I
- Spring 2010 Actuarial Mathematics I
- Fall 2009 Actuarial Mathematics III
- Spring 2010 Actuarial Mathematics III
Mentoring before Solis-Lemus lab
For members of Solis-Lemus lab, see Team
2019
- Software Development Project, Fast reconstruction and visualization of phylogenetic networks, Harnoor Singh and Naman Kanwar – Georgia State University.
2018
- Software Development Project, Julia package for GAMuT test, Anna Voss - Emory University
- Statistics Senior Honors Thesis, GWAS data analysis for epilepsy, Mengtong Hu - Emory University
2016
- Software Development Project, Parallelization of SNaQ in PhyloNetworks, Josh McGrath - University of Wisconsin-Madison
- Undergraduate Research Scholar, Computing tools for bayesian phylogenetic inference, Jordan Vonderwell - University of Wisconsin-Madison
- Statistics Senior Honors Thesis, Computing tools and performance of PhyloNetworks, Nan Ji - University of Wisconsin-Madison
2015
- Statistics Senior Honors Thesis, Inconsistency of species-tree methods under gene flow, Mengyao Yang – University of Wisconsin-Madison
- Integrated Biological Sciences Summer Research Program, Visualizing inferred phylogenetic networks in Julia, John Spaw – University of Wisconsin-Madison
2014
- Integrated Biological Sciences Summer Research Program, Using quartets to estimate phylogenetic networks, John Malloy – University of Wisconsin-Madison.